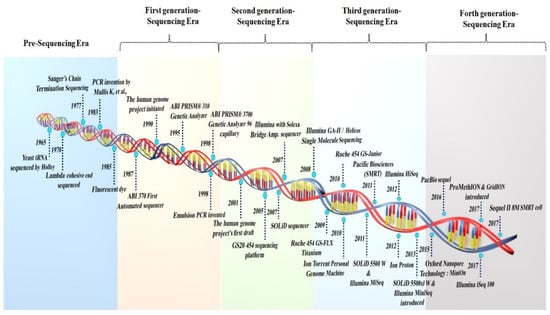
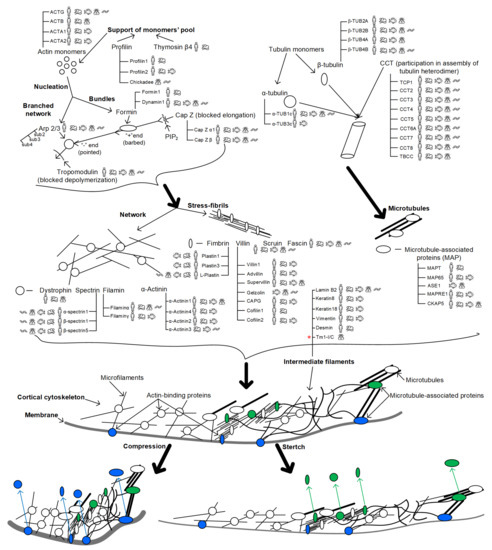
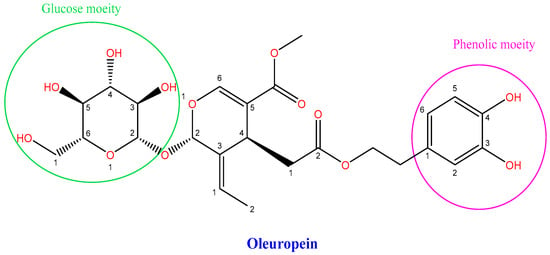
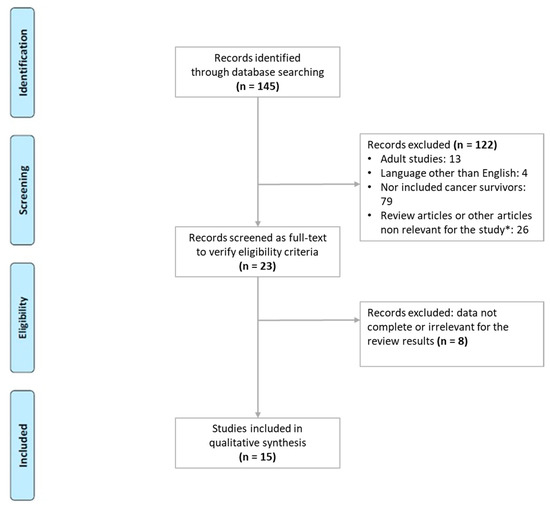
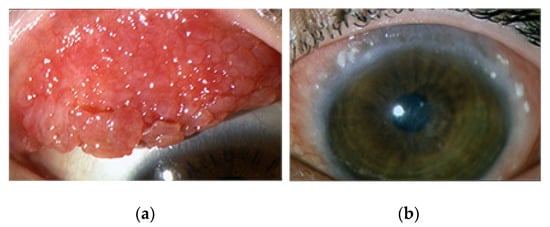
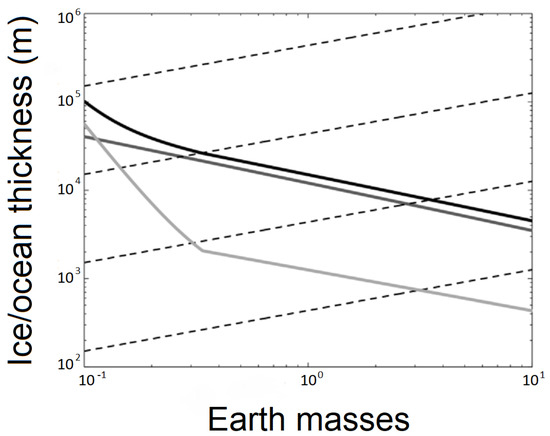
<p>Aquaporin structure. (<b>a</b>) AQP side monomer view parallel to the membrane (the two NPA motifs are shown in yellow), (<b>b</b>) top view of the AQP tetramer from the extracellular side (water molecules in the pore are shown as red spheres), (<b>c</b>) the AQP monomer with the six transmembrane alpha-helices (1–6) connected by five loops extending into the extracellular and cytoplasmic spaces (A–E), and (<b>d</b>) illustration of the AQP hourglass-shaped channel, depicting the two selectivity filters (ar/R and NPA), allowing the passage of water molecules and other solutes, as shown, while excluding ions and larger solutes. Abbreviations: AQP, aquaporin; ar/R, aromatic/arginine; NPA, asparagine-proline-alanine. Image adapted from [<a href="#B33-life-14-01688" class="html-bibr">33</a>,<a href="#B83-life-14-01688" class="html-bibr">83</a>]. Under Creative Commons CC BY 4.0 license.</p> Full article ">Figure 2
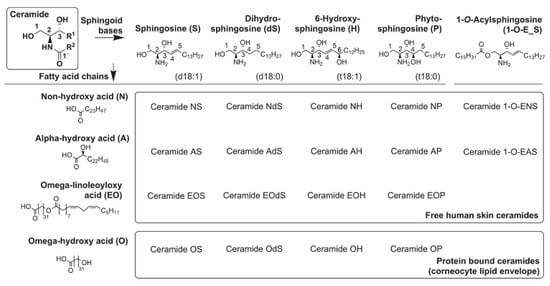
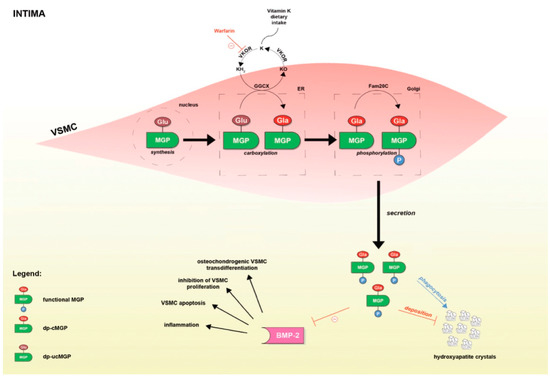
<p>Regulation of aquaporin expression and function. Aquaporin gene expression is tightly regulated by several factors including cytokine levels, hypoxia, cell osmotic conditions, and vasopressin. Following RNA expression, alternative splicing events and regulatory molecules such as micro RNAs (miRNAs), act upon the RNA molecules, thus regulating protein production. A series of post-translational modifications, including phosphorylation, methylation, and ubiquitination determine the function and localization of the formed proteins. AQPs may be alternatively stored in intracellular vesicles to be rapidly transported to the cell membrane upon stimulation. Abbreviations: AQP, aquaporin; miRNAs, micro RNAs.</p> Full article ">Figure 3
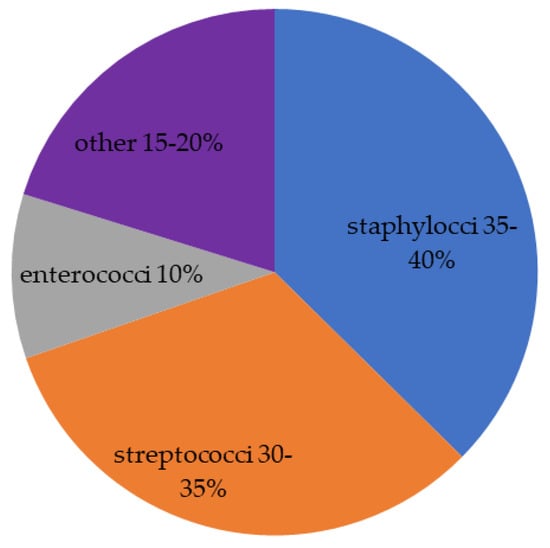
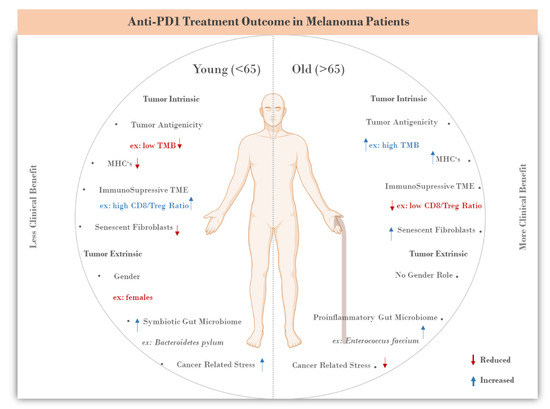
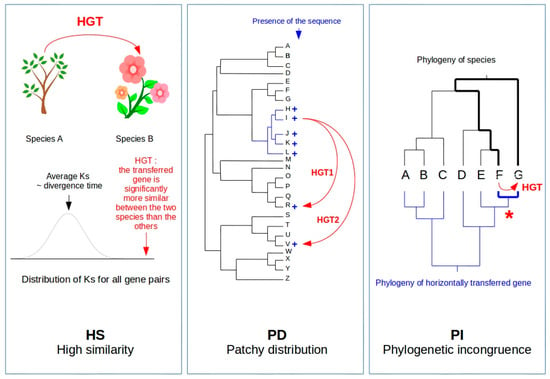
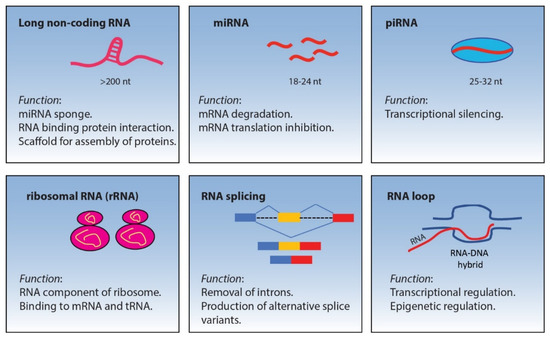
<p>Aquaporin expression in human immune cells and their proposed roles in sepsis. AQP1 is expressed in neutrophils, monocytes, and lymphocytes, contributing to water transport and cell volume regulation during migration and differentiation. AQP3 is expressed in macrophages, lymphocytes, and dendritic cells, facilitating chemokine-driven migration and supporting energy metabolism through glycerol transport. AQP5 is expressed in some immune cells, particularly lymphocytes, neutrophils, and dendritic cells. AQP5 plays a role in facilitating water transport, which is critical for maintaining cell shape and motility during migration. AQP9 is most prominently expressed in leukocytes, aiding in respiratory bursts and cell motility during pathogen defense. It is expressed in neutrophils, macrophages, monocytes, lymphocytes, and dendritic cells. Other AQPs detected at low concentrations in immune cells are AQP6, AQP7, AQP8, AQP11, and AQP12, expressed in monocytes, macrophages, and dendritic cells. AQPs are important regulators of fluid balance/edema, inflammation/immune response, and vascular permeability. During inflammatory conditions, such as sepsis, AQP expression is altered and correlates with disease severity and outcomes. Abbreviations: AQP, aquaporin; ROS, reactive oxygen species. Image adapted from [<a href="#B117-life-14-01688" class="html-bibr">117</a>].</p> Full article ">Figure 4
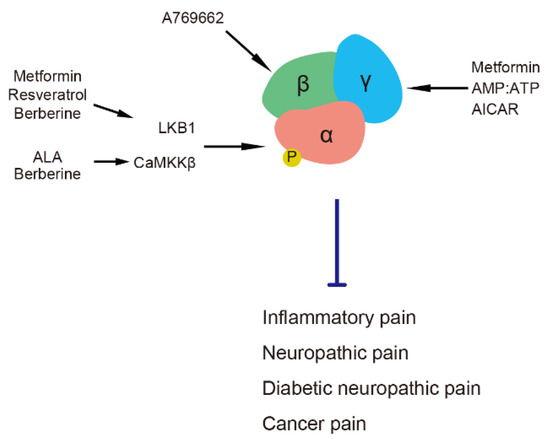
<p>Aquaporins in the human lung. The lung facilitates oxygen and carbon dioxide exchange between air and blood. Maintaining the fluid volume is essential for gas exchange, achieved by balancing fluid transport across the epithelium. Aquaporins (AQPs) are integral membrane proteins that act as water channels, facilitating water transport across cell membranes in response to osmotic gradients. Under physiological conditions, aquaporins usually facilitate fluid management by transporting it out of the alveolar space into the interstitium or capillaries. Τhe main aquaporins expressed in the lung are AQP1, AQP3, AQP4, and AQP5. AQP1 is present in the capillary endothelial cells and controls water movement across the endothelial barrier. AQP3 and AQP4 are found in the airway epithelium and alveolar epithelial cells. AQP5 is expressed in the alveolar epithelial cells type I and is primarily responsible for water transport across the alveolar barrier. Acute respiratory distress syndrome (ARDS) constitutes a severe lung condition characterized by the widespread release of pro- and anti-inflammatory cytokines, oxidative stress (production of reactive oxygen species-ROS), increased vascular permeability, and accumulation of fluid in the alveoli, which leads to impaired gas exchange and respiratory failure. In ARDS, increased water transport by AQPs could contribute to fluid accumulation in the alveoli if the inflammation and barrier dysfunction are uncontrolled, exacerbating pulmonary edema. Disruption of the expression or function of these AQPs, as is often observed in ARDS, compromises the lung’s ability to clear fluid, leading to worsened pulmonary edema. Abbreviations: AQP, aquaporin; ROS, reactive oxygen species. Adapted from [<a href="#B157-life-14-01688" class="html-bibr">157</a>].</p> Full article ">Figure 5
<p>Aquaporins in the human kidney. Aquaporins (AQPs) play a critical role in kidney function by regulating water transport across membranes, crucial for urine concentration and maintaining fluid balance. AQP1 facilitates water reabsorption in the proximal tubules and descending limb of the loop of Henle, while AQP2 is key in the collecting ducts, regulated by antidiuretic hormone (ADH) to concentrate urine. AQPs 3 and 4 support water exit from the collecting duct cells to the bloodstream. AQP7 is expressed in the brush border of the S3 segment of the proximal tubule and plays a significant role in metabolism by regulating glycerol transport. AQP8 enables bidirectional water and hydrogen peroxide transport across biological membranes, while AQP11 is uniquely located in the endoplasmic reticulum membrane of proximal tubular cells. Dysfunction of AQPs can lead to conditions like nephrogenic diabetes insipidus or fluid imbalances. Panels on the left show proposed mechanisms of aquaporin involvement in AKI. The panel on the right shows approaches that could regulate AQP expression. The downward arrows designate decreased expression. Abbreviations: AKI, acute kidney injury; AQP, aquaporin; H<sub>2</sub>O<sub>2</sub>, hydrogen peroxide; I/R, ischemia/reperfusion; LPS, lipopolysaccharide. Image reproduced from [<a href="#B24-life-14-01688" class="html-bibr">24</a>]. Aspects of this image were partially adapted. Under Creative Commons CC BY 4.0 license.</p> Full article ">Figure 6
<p>Main pathophysiological roles of aquaporins in acute brain injury. Aquaporins (AQPs) are pivotal in the pathophysiology of acute brain injury, and their role extends across key processes such as cerebral edema formation, blood–brain barrier regulation, and inflammatory response modulation. AQP4 has emerged as a central player, influencing water homeostasis and edema resolution, while other aquaporins, such as AQP1, AQP2, and AQP9, contribute to diverse pathophysiological mechanisms, including immune cell migration and metabolic adaptation. Aspects of this figure were adapted with permission from Servier Medical Art library, available under Creative Commons license.</p> Full article ">