1 Introduction
The majority of dynamic Web content is backed by relational databases (RDB), and so are many enterprise systems [DynaWebSites]. On the other hand, in order to expose structured data on the Web, Resource Description Framework (RDF) [RDF] is used. This document reviews use cases and requirements for a relational database to RDF mapping (RDB2RDF) with the following structure:
- The remainder of this section motivates why mapping RDBs to RDF is necessary and needed and highlights the importance of a standard.
- In the next section RDB2RDF use cases are reviewed.
- The last section discusses requirements regarding a RDB2RDF mapping language, driven by an analysis of the aforementioned use cases.
1.1 Why Mapping RDBs to RDF?
The Web of Data is constantly growing due to its compelling potential of facilitating data integration and retrieval. At the same time however, RDB systems host a vast amount of structured data in relational tables augmented with integrity constraints. In order to make this huge amount of relational data available for the Web of Data, a connection must be established between RDBs and a format suitable for the Web of Data.
The advantages of creating an RDF view of relational data are inherited from the Web of Data and can be summarized based on the tasks they facilitate:
-
Integration: data in different RDBs can be integrated using RDF semantics and mechanisms; in this sense, the Web of Data can be imagined as one big database. Moreover, information in the database can be integrated with information that comes from other data sources.
-
Retrieval: once data are published in the Web of Data (as opposed to relational databases), queries can span different data sources and more powerful retrieval methods can be built.
RDF data in the Web should be defined and linked in a way that makes it accessible for humans and machines [LinkedData]. The Web of data is a scalable environment with explicit semantics where not only humans can navigate information, but also machines are able to find connections and use them to navigate through the information space. In order to realise this global information space, we need to:
- follow a common model to describe, connect and access resources;
- name resources in an unambiguous way
The most common way to publish resources in the Web of Data follows the RDF model [RDF] and uses Uniform Resource Identifiers ([URI]) for resource identification, thereby facilitating the creation of a comprehensive and flexible resource description.
In the following, we will use RDB2RDF to denote any technique that takes as an input a RDB (schema and data) and produces one or more RDF graphs, as depicted in the following figure:
The consumer of the RDF Graph (virtual or materialized) essentially can access the RDF data in different ways:
-
Query access, which means the agent issues a SPARQL query against an endpoint exposed by the system and processes the results (typically the result is a SPARQL result set in XML or JSON);
-
Entity-level access, which means the agent performs an HTTP
GET on a URI exposed by the system and processes the result (typically the result is an RDF graph);
-
Dump access, which means the agent performs an HTTP
GET on dump of the entire RDF graph, for example in Extract, Transform, and Load (ETL) processes.
1.2 Why a standard RDB2RDF method?
With the advent of the Web of Data, researcher and practitioners have compared the RDB model and the RDF model [RDB-RDF] and surveyed different approaches to map them [RDBMSMapping]. As noted in the survey on existing RDB2RDF mapping approaches [RDB2RDFSurvey], a couple of proposals on how to tackle the RDB2RDF mapping issues are known.
Use of a standard for mapping language for RDB to RDF may allow use of a single mapping specification in the context of mirroring of schema and (possibly some or all of the) data in various databases, possibly from different vendors (e.g., Oracle database, MySQL, etc.) and located at various sites. Similarly structured data (that is, data stored using same schema) is useful in many different organizations often located in different parts of the world. These organizations may employ databases from different vendors due to one or more of many possible factors (such as, licensing cost, resource constraints, availability of useful tools and applications and of appropriate database administrators, etc.). Presence of a standard RDB2RDF mapping language allows creation and use of a single mapping specification against each of the hosting databases to present a single (virtual or materialized) RDF view of the relational data hosted in those databases and this RDF view can then be queried by applications using SPARQL query or protocol.
Another reason for a standard is to allow easy migration between different systems. Just as a single web-page in HTML can be viewed by two different Web browsers from different vendors, a single RDB2RDF mapping standard should allow a user from one database to expose their data as RDF, and then, when they export their data to another database, allow the newly imported data to be queried as RDF without changing the mapping file. For example, imagine that a database administrator is exposing weather data as Linked Data to be consumed by other applications. At first, this weather data is stored in a light-weight database (such as MySQL). However, as more and more weather data is collected, and more and more users access the Web data, the light-weight database may have difficulty scaling. Therefore, the database administrator migrates their database to a more heavy-weight database (such as Oracle Database 11g). Of course, the database administrator does not want to re-create the ability to view the data as RDF using a vendor-specific mapping file, but instead wants to seamlessly migrate the view of their data as RDF.
A standardized mapping between relational data and RDF allows the database administrator to migrate the view of their data as RDF across databases, allowing the vendors to compete on functionality and features rather than forcing database administrators to rewrite their entire relational data to RDF mapping when they want to migrate their data from one database to another.
Another motivation for a standard is that for certain classes of systems (such as CMS) a 'default' mapping could be defined which can be deployed no matter what underlying RDB is used. As these systems, such as Drupal or Wordpress, can be run on top of different underlying relational databases. A standardized way of mapping between relational data and RDF hence allows the underlying database to be changed without disturbing the content management system.
Further, having a standard mapping would simplify programming applications that access multiple database sources.
2 Use Cases
With integration of relational data from one RDB with various kinds of data (such as relational, spreadsheets, CSV, unstructured text, etc.), the use cases presented in the following fall into one or more of the following categories:
- I want to integrate my RDB with another structured source (RDB, XLS, CSV, etc.), so I'll convert my RDB to RDF and assume my other structured source can also be in RDF. See UC1, UC3.
- I want to integrate my RDB with existing RDF on the web (Linked Data), so I'll convert my RDB to RDF and then I'm able to link and integrate. See UC2, UC4.
- I want to integrate my RDB with unstructured data (HTML, PDF, etc.), so I'll convert my RDB to RDF and assume my other unstructured source can also be in RDF.
- I want my RDB data to be available for SPARQL or other RDF-based querying, and/or for others to integrate with other data sources (structured, RDF, unstructured). See UC2, UC4.
2.1 UC1 - Patient Recruitment
The Semantic Web for Health Care and Life Sciences Interest Group has created several demonstrators using SPARQL to query clinical and biological relational databases. Included is the database structure, sample data, and a SPARQL query. Following are six tables of sample diabetic patient data extracted from the University of Texas Health Science Center. Some columns have been omitted from this use case for brevity.
Accompanying each table are two RDF views (represented in Turtle) corresponding to HL7/RIM and CDISK SDTM ontology in RDFS. While there are many motivations for providing a common interface to administer distinct databases (access to patient history, shared rules for clinical decision support, etc.), in this case, SPARQL queries (following the table description) were used to find candidates for clinical studies. For these RDF graphs, the following namespaces apply:
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .
@prefix hl7: <http://www.hl7.org/v3ballot/xml/infrastructure/vocabulary/vocabulary#> .
@prefix stdm: <http://www.sdtm.org/vocabulary#> .
Person
|
ID
|
SexDE
|
DateOfBirth
|
LastEditedDTTM
|
|
1234561
|
2
|
1983-01-02 00:00:00
|
2007-11-13 15:49:20
|
|
1234562
|
3
|
1963-12-27 00:00:00
|
2008-01-30 17:08:42
|
|
1234563
|
2
|
1983-02-25 00:00:00
|
2007-03-10 06:01:55
|
<http://hospital.example/DB/Person/ID.1234561#record> a hl7:Person ;
hl7:administrativeGenderCodePrintName Sex_DE:M ;
hl7:livingSubjectBirthTime "1983-01-02T00:00:00Z"^^xsd:dateTime .
<http://hospital.example/DB/Person/ID.1234562#record> a hl7:Person ;
hl7:administrativeGenderCodePrintName Sex_DE:F ;
hl7:livingSubjectBirthTime "1963-12-27T00:00:00Z"^^xsd:dateTime .
<http://hospital.example/DB/Person/ID.1234563#record> a hl7:Person ;
hl7:administrativeGenderCodePrintName Sex_DE:M ;
hl7:livingSubjectBirthTime "1983-02-25T00:00:00Z"^^xsd:dateTime .
<http://hospital.example/DB/Person/ID.1234561#record> a sdtm:Patient ;
stdm:dateTimeOfBirth "1983-01-02T00:00:00Z"^^xsd:dateTime .
<http://hospital.example/DB/Person/ID.1234562#record> a sdtm:Patient ;
sdtm:dateTimeOfBirth "1963-12-27T00:00:00Z"^^xsd:dateTime ;
sdtm:sex <http://hospital.example/DB/Sex_DE#F> .
<http://hospital.example/DB/Person/ID.1234563#record> a sdtm:Patient ;
sdtm:dateTimeOfBirth "1983-02-25T00:00:00Z"^^xsd:dateTime ;
sdtm:sex <http://hospital.example/DB/Sex_DE#F> .
Sex_DE
|
ID
|
EntryCode
|
EntryName
|
EntryMnemonic
|
|
2
|
1
|
Male
|
M
|
|
3
|
2
|
Female
|
F
|
<http://hospital.example/DB/Zex_DE/ID.2#record>
hl7:administrativeGenderCodePrintName "Male"@en-us ;
Sex_DE:EntryMnemonic "M"@en-us .
<http://hospital.example/DB/Zex_DE/ID.3#record>
hl7:administrativeGenderCodePrintName "Female"@en-us ;
Sex_DE:EntryMnemonic "F"@en-us .
<http://hospital.example/DB/Person/ID.1234561#record>
stdm:sex <http://hospital.example/DB/Sex_DE#M> .
<http://hospital.example/DB/Person/ID.1234562#record>
stdm:sex <http://hospital.example/DB/Sex_DE#F> .
<http://hospital.example/DB/Person/ID.1234563#record>
stdm:sex <http://hospital.example/DB/Sex_DE#M> .
Item_Medication
|
ID
|
PatientID
|
ItemType
|
PerformedDTTM
|
|
99999999002
|
1234561
|
ME
|
2007-09-28 00:00:00
|
|
99999999003
|
1234562
|
ME
|
2007-09-28 00:00:00
|
|
99999999004
|
1234562
|
ME
|
2008-07-28 00:00:00
|
<http://hospital.example/DB/Person/ID.1234561#record>
hl7:substanceAdministration <http://hospital.example/DB/Item_Medication/ID.99999999002#record> .
<http://hospital.example/DB/Item_Medication/ID.99999999002#record>
a hl7:SubstanceAdministration ;
hl7:effectiveTime _:t1 .
_:t1 hl7:start "2007-09-28T00:00:00"^^xsd:dateTime .
<http://hospital.example/DB/Person/ID.1234562#record>
hl7:substanceAdministration <http://hospital.example/DB/Item_Medication/ID.99999999003#record> ;
hl7:substanceAdministration <http://hospital.example/DB/Item_Medication/ID.99999999004#record> .
<http://hospital.example/DB/Item_Medication/ID.99999999003#record>
a hl7:SubstanceAdministration ;
hl7:effectiveTime _:t2 .
_:t2 hl7:start "2007-09-28T00:00:00"^^xsd:dateTime .
<http://hospital.example/DB/Item_Medication/ID.99999999004#record>
a hl7:SubstanceAdministration ;
hl7:effectiveTime _:t3 .
_:t3 hl7:start "2008-07-28T00:00:00"^^xsd:dateTime .
Medication
|
ID
|
ItemID
|
Dose
|
Refill
|
QuantityToDispense
|
DaysToTake
|
PrescribedByID
|
MedDictDE
|
|
88888888002
|
99999999002
|
2
|
6
|
180
|
45
|
1004682
|
132139
|
|
88888888003
|
99999999002
|
2
|
0
|
180
|
45
|
1004683
|
132139
|
|
88888888004
|
99999999003
|
2
|
6
|
180
|
45
|
1004682
|
132139
|
|
88888888005
|
99999999004
|
4
|
6
|
180
|
45
|
1004682
|
132139
|
<http://hospital.example/DB/Item_Medication/ID.99999999002#record>
hl7:consumable <http://hospital.example/DB/Medication_DE/ID.132139#record> .
_:t1 hl7:durationInDays 45 .
<http://hospital.example/DB/Item_Medication/ID.99999999003#record>
hl7:consumable <http://hospital.example/DB/Medication_DE/ID.132139#record> .
<http://hospital.example/DB/Item_Medication/ID.99999999004#record>
hl7:consumable <http://hospital.example/DB/Medication_DE/ID.132139#record> .
_:t2 hl7:durationInDays 45 .
<http://hospital.example/DB/Item_Medication/ID.99999999005#record>
hl7:consumable <http://hospital.example/DB/Medication_DE/ID.132139#record> .
_:t3 hl7:durationInDays 45 .
Medication_DE
|
ID
|
Entry
|
EntryCode
|
EntryName
|
NDC
|
Strength
|
Form
|
UnitOfMeasure
|
DrugName
|
DisplayName
|
|
132139
|
131933
|
98630
|
GlipiZIDE-Metformin HCl 2.5-250 MG Tablet
|
54868079500
|
2.5-250
|
TABS
|
MG
|
GlipiZIDE-Metformin HCl
|
GlipiZIDE-Metformin HCl 2.5-250 MG Tablet
|
<http://hospital.example/DB/Medication_DE/ID.132139#record>
hl7:displayName "GlipiZIDE-Metformin HCl 2.5-250 MG Tablet" .
NDCcodes
|
ingredient
|
RxCUI
|
labelType
|
name
|
NDC
|
|
6809
|
351273
|
Clinical
|
Glipizide 2.5 MG / Metformin 500 MG Oral Tablet
|
54868079500
|
<http://hospital.example/DB/Medication_DE/ID.132139#record>
spl:activeIngredient _:i1 .
_:i1 spl:classCode 54868079500 .
[ a sdtm:ConcomitantMedication ;
sdtm:subject <http://hospital.example/DB/Person/ID.1234561#record> ;
sdtm:standardizedMedicationName "GlipiZIDE-Metformin HCl 2.5-250 MG Tablet" ;
hl7:activeIngredient [hl7:classCode 54868079500 ] ;
sdtm:startDateTimeOfMedication "2007-09-28 00:00:00"^^xsd:dateTime ] .
[ a sdtm:ConcomitantMedication ;
sdtm:subject <http://hospital.example/DB/Person/ID.1234562#record> ;
sdtm:standardizedMedicationName "GlipiZIDE-Metformin HCl 2.5-250 MG Tablet" ;
hl7:activeIngredient [ hl7:classCode 54868079500 ] ;
sdtm:startDateTimeOfMedication "2007-09-28 00:00:00"^^xsd:dateTime ] .
[ a sdtm:ConcomitantMedication ;
sdtm:subject <http://hospital.example/DB/Person/ID.1234562#record> ;
sdtm:standardizedMedicationName "GlipiZIDE-Metformin HCl 2.5-250 MG Tablet" ;
hl7:activeIngredient [ hl7:classCode 54868079500 ] ;
sdtm:startDateTimeOfMedication "2008-07-28 00:00:00"^^xsd:dateTime ] .
2.1.1 Queries Over the RDF Graph
The use case can be realised as a SPARQL query over the RDF graphs. The following is a SPARQL query, which extracts patients taking a particular class of medication, an anticoagulant, and not another (weight loss, here).
PREFIX sdtm: <http://www.sdtm.org/vocabulary#>
PREFIX spl: <http://www.hl7.org/v3ballot/xml/infrastructure/vocabulary/vocabulary#>
SELECT ?patient ?dob ?sex # ?takes ?indicDate ?indicEnd ?contra
WHERE {
?patient a sdtm:Patient ;
sdtm:middleName ?middleName ;
sdtm:dateTimeOfBirth ?dob ;
sdtm:sex ?sex .
[ sdtm:subject ?patient ;
sdtm:standardizedMedicationName ?takes ;
spl:activeIngredient [ spl:classCode 6809 ] ;
sdtm:startDateTimeOfMedication ?indicDate
] .
OPTIONAL {
[ sdtm:subject ?patient ;
sdtm:standardizedMedicationName ?disqual ;
spl:activeIngredient [ spl:classCode 11289 ]
sdtm:startDateTimeOfMedication ?indicDate
] }
} LIMIT 30
2.1.2 Derived Requirements
This use case leads to the following requirements:
- Map relational data to an RDF schema derived from the relational schema.
- Create a virtual RDF view that is used to translate SPARQL queries over the RDF views to SQL queries over the relational data.
- Mapping of relational data types to RDF/XML datatypes, for example, SQL date/time formats to XML Schema date/time formats.
- Mapping column names to RDF property names, for example, the column
DaysToTake from the Medication table is mapped to hl7:durationInDays.
2.2 UC2 - Web applications (Wordpress)
In order to make the Web of Data useful to ordinary Web users, RDF and OWL have to be deployed on the Web on a much larger scale. Web applications such as Content Management Systems, online shops or community applications (e.g. Wikis, blogs, forums) already store their data in relational databases. Providing a standardized way to map the relational data and schema behind these Web applications into RDF, RDF-Schema and OWL will facilitate novel semantic browsing and search applications.
By supporting the long tail of Web applications and thus counteracting the centralization of the Web 2.0 applications, the planned RDB2RDF standardization will help to give control over data back to end-users and thus promote a democratization of the Web.
To support this use case, the mapping language should be easily implementable for lightweight Web applications and have a shallow learning curve to foster early adoption by Web developers.
We illustrate this use case with the example of Wordpress. Wordpress is a popular blogging Web application and installed on tens of thousands of Web servers. Wordpress used a relational database (MySQL) with a relatively simple schema:
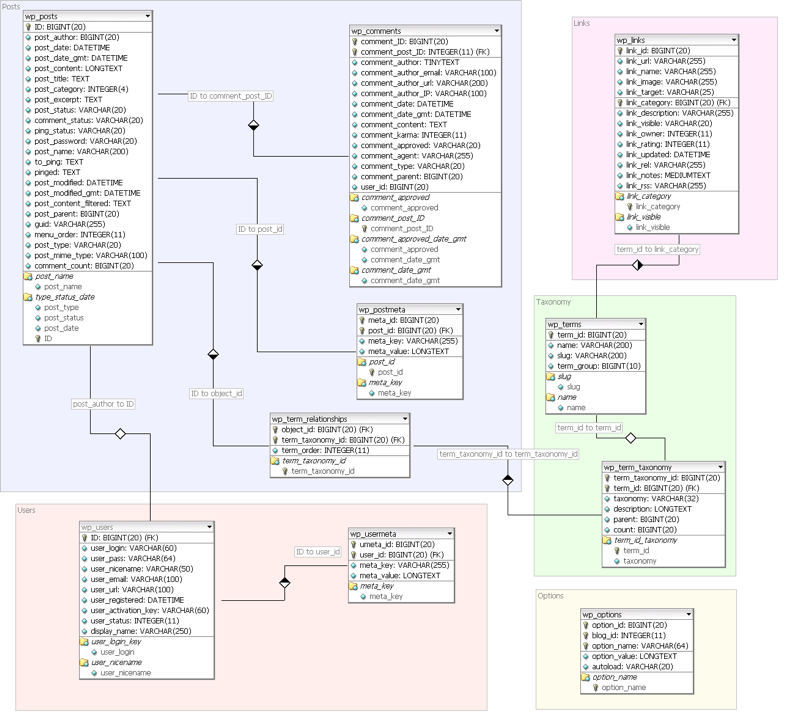
Wordpress SQL Schema:
A mapping should be able to reuse existing vocabularies.
Mapped to RDF the resulting ontology should contain the classes post, attachment, tag, category, user and comment. An example instance of the post class, for example, should look like:
@prefix sioc: <http://rdfs.org/sioc/ns#> .
@prefix dc: <http://purl.org/dc/terms/> .
@prefix dc11: <http://purl.org/dc/elements/1.1/> .
<http://blog.aksw.org/triplify/post/8> a sioc:Post .
<http://blog.aksw.org/triplify/post/8> sioc:has_creator <http://blog.aksw.org/triplify/user/5> .
<http://blog.aksw.org/triplify/post/8> dc:created "2007-02-27T17:23:36"^^<http://www.w3.org/2001/XMLSchema#dateTime> .
<http://blog.aksw.org/triplify/post/8> dc11:title "Submissions open: 3rd Workshop on Scripting for the Semantic Web" .
<http://blog.aksw.org/triplify/post/8> sioc:content "The submissions web-site is now open for the 3rd Workshop on Scripting for the Semantic Web..." .
<http://blog.aksw.org/triplify/post/8> dc:modified "2008-02-22T21:41:00"^^<http://www.w3.org/2001/XMLSchema#dateTime> .
2.2.1 Queries Over the RDF Graph
An example SPARQL query on the resulting ontology could be:
SELECT ?name, ?title
WHERE {
?post rdf:type sioc:Post .
?post dc:title ?title .
?post dc:has_creator ?author
?author foaf:name ?name
}
2.2.2 Derived Requirements
This use case leads to the following requirements:
- Support Extract-Transform-Load (ETL) for the created RDF by the mapping.
2.3 UC3 - Integrating Enterprise Relational databases for tax control
Integrating relational databases and exposing them on the Web or Intranet requires the re-use of unique identifiers in order to integrate and interlink data about entities on different databases.
The re-use of unique identifiers allows:
- Joins between entities described in different databases.
- Join structured data (SQL) to structured data, from incompatible schema, or where data is dirty, poorly normalized, lacking proper keys/indices.
This use case is a pilot project for the Trentino region tax agency. Trentino is an autonomous region in the north of Italy. The region has a population of 1 million people and more than 200 municipalities with their own information systems. The goal of is to integrate and link tax related data about people, organizations, buildings, etc. This data come from different databases especially from the region's many municipalities, each with their own individual data structures.
The re-use of unique identifiers will provide a lightweight method for aggregating the data. In this way we are providing a tax agent an intelligent tool for navigating through the data present in the many different databases. Using unique identifiers, a tool can aggregate data and create a profile for each tax payer. Each user profile shows different type of information, with links to other entities such as the buildings owned, payments made, location of residence etc.
The RDF generated from the two databases is materialized and joined using the generated unique identifiers.
Example
Supposed that we have two tables (Anagrafe and Urban_Cadastre) from different databases, we select some typical attributes for the two tables to explain our conversion method. Table Angrafe includes the information about two type of entities, persons and locations (a person's residence place), and some other information:
|
Firstname
|
Lastname
|
City_Residence_Place
|
Country_Residence_Place
|
Other_Info
|
|
Paolo
|
Bouquet
|
Trento
|
Italy
|
xyz...
|
Table Urban_Cadastre contains the information about buildings and their owners:
|
Owner_Name
|
Building_LocalID
|
Building_Address
|
Building_Type
|
|
Paolo Bouquet
|
123456
|
VIA G.LEOPARDI
|
3
|
DDL:
CREATE TABLE Anagrafe (
Firstname varchar
Lastname varchar
City_Residence_Place varchar
Country_Residence_Place varchar
Other_Info varchar
PRIMARY KEY (Firstname, Lastname)
)
CREATE TABLE (
Owner_Name varchar
Building_LocalID integer
Building_Address varchar
Building_Type varchar
PRIMARY KEY (Owner_Name)
)
Using traditional RDB2RDF translation methods the RDF representation for the two example tables coming from two different databases is shown below:
@prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> .
@prefix database_anagrafe: <http://www.database1.org/anagrafe/> .
database_anagrafe:entry_row1 database_anagrafe:Other_Info "xyz" ;
database_anagrafe:Country_Residence_Place "Italy" ;
database_anagrafe:City_Residence_Place "Trento" ;
database_anagrafe:Last_Name "Bouquet" ;
database_anagrafe:First_Name "Paolo" .
@prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> .
@prefix database_urbano: <http://www.database1.org/Urban_Cadastre/> .
database_urbano:entry_row1 database_urbano:Building_Type "3" ;
database_urbano:Building_Address "VIA G.LEOPARDI" ;
database_urbano:Building_LocalID "123456" ;
database_urbano:Owner_Name "Paolo Bouquet" .
2.3.1 Querying
If we wanted to query these two tables, we would have to create a unique identifier, such as http://www.example.org/Paolo_Bouquet for Paolo Bouquet, to refer to entities in order to join descriptions about the same entity coming from different data sources.
2.3.2 Derived Requirements
This use case leads to the following requirements:
- Create unique identifiers for the entities described by the data.
2.4 UC4 - rCAD: RNA Comparative Analysis Database
rCAD - RNA Comparative Analysis using SQLServer: the tremendous increase in available biological information creates opportunities to decipher the structure, function and evolution of cellular components while presenting new computational challenges for performance and scalability. To fully utilize this large increase in knowledge, it must be organized for efficient retrieval and integrated for multi-dimensional analysis. Given this, biologists are able to invent new comparative sequence analysis protocols that will yield new and different structural and functional information. Based on Microsoft SQL-server, we have designed and implemented the RNA Comparative Analysis Database - rCAD which supports comparative analysis of RNA sequence and structure, and unites, for the first time in a single environment, multiple dimensions of information necessary for alignment viewing, sequence metadata, structural annotations, structure prediction studies, structural statistics of different motifs, and phylogenetic analysis. This system provides a queryable environment that hosts efficient updates and rich analytics.
For this use-case, we will be presenting only the Sequence Alignment schema. The SQL-DDL for Microsoft SQL Server can be found here: rCAD Sequence Alignment SQL DDL
This use-case presents the issue of a schema that does not have an existing domain ontology in which it can be mapped to. The closest domain ontology is the Multiple Alignment Ontology (MAO) which can only be mapped to the Sequence Alignment part of the entire rCAD database. However, MAO is in the OBO language. Nevertheless, OBO ontologies can be translated to OWL (and back).
2.4.1 Expose Relational Data as RDF when there is no existing Domain Ontology to map the relational schema to
Rob from the RNA lab would like to expose the Sequence Alignment data from the rCAD database as RDF. However, there is no existing domain ontology in which the relational schema can be mapped to. Therefore, an ontology should be derived automatically from the relational schema. The RDF data will become instance of this automatically generated ontology.
The Alignment table from the rCAD database is the following:
CREATE TABLE [AlignmentClassic].[Alignment] (
[AlnID] int NOT NULL,
[SeqTypeID] tinyint NOT NULL,
[AlignmentName] varchar(max) NULL,
[ParentAlnID] int NULL,
[NextColumnNumber] int NOT NULL,
CONSTRAINT [PK_Alignment] PRIMARY KEY([AlnID])
)
ON [PRIMARY]
GO
ALTER TABLE [AlignmentClassic].[Alignment]
ADD CONSTRAINT [FK_Alignment_SequenceType]
FOREIGN KEY([SeqTypeID])
REFERENCES [dbo].[SequenceType]([SeqTypeID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
ALTER TABLE [AlignmentClassic].[Alignment]
ADD CONSTRAINT [FK_Alignment_Alignment]
FOREIGN KEY([ParentAlnID])
REFERENCES [AlignmentClassic].[Alignment]([AlnID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
and the desired ontology that is generated automatically from the relational schema is the following:
PREFIX rcad: <http://rcad.org/vocabulary/rcad.owl#>
rcad:Alignment rdf:type owl:Class .
rcad:AlignmentName rdf:type owl:DatatypeProperty.
rcad:AlignmentName rdfs:domain rcad:Alignment.
rcad:AlignmentName rdfs:range xsd:string.
rcad:NextColumnNumber rdf:type owl:DatatypeProperty.
rcad:NextColumnNumber rdfs:domain rcad:Alignment.
rcad:NextColumnNumber rdfs:range xsd:integer.
rcad:ParentAlnID rdf:type owl:ObjectProperty
rcad:ParentAlnID rdfs:domain rcad:Alignment
rcad:ParentAlnID rdfs:range rcad:Alignment
and the desired RDF triples, which are instances of the automatically generated ontology are the following:
PREFIX rcad: <http://rcad.org/vocabulary/rcad.owl#>
PREFIX rcad-data: <http://rcad.org/vocabulary/rcad-data.rdf#>
rcad-data:alignment1000 a rcad:Alignment;
rcad:AlignmentName "My Alignment 1000"^^xsd:string;
rcad:NextColumnNumber "123"^^xsd:int;
rcad:ParentAlnID rcad-data:alignment2000
2.4.2 Expose Mapping a Relational Database to an OWL DL ontology
Rob, from the RNA lab would like to expose the Sequence Alignment data from the rCAD database as RDF. Just recently the Multiple Alignment Ontology (MAO) as been released, which is an "ontology for data retrieval and exchange in the fields of multiple DNA/RNA alignment, protein sequence and protein structure alignment." However, this ontology has been developed in OBO. Nevertheless, OBO ontologies can be translated to OWL ontologies, specifically OWL DL. Therefore, Rob will like to map his rCAD database to the MAO ontology
The Alignment and Alignment Column tables from the rCAD database is the following:
CREATE TABLE [AlignmentClassic].[Alignment] (
[AlnID] int NOT NULL,
[SeqTypeID] tinyint NOT NULL,
[AlignmentName] varchar(max) NULL,
[ParentAlnID] int NULL,
[NextColumnNumber] int NOT NULL,
CONSTRAINT [PK_Alignment] PRIMARY KEY([AlnID])
)
GO
ALTER TABLE [AlignmentClassic].[Alignment]
ADD CONSTRAINT [FK_Alignment_SequenceType]
FOREIGN KEY([SeqTypeID])
REFERENCES [dbo].[SequenceType]([SeqTypeID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
ALTER TABLE [AlignmentClassic].[Alignment]
ADD CONSTRAINT [FK_Alignment_Alignment]
FOREIGN KEY([ParentAlnID])
REFERENCES [AlignmentClassic].[Alignment]([AlnID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
---
CREATE TABLE [AlignmentClassic].[AlignmentColumn] (
[AlnID] int NOT NULL,
[ColumnNumber] int NOT NULL,
[ColumnOrdinal] int NOT NULL,
CONSTRAINT [PK_AlignmentColumn] PRIMARY KEY([AlnID],[ColumnNumber])
)
GO
ALTER TABLE [AlignmentClassic].[AlignmentColumn]
ADD CONSTRAINT [FK_AlignmentColumn_Alignment]
FOREIGN KEY([AlnID])
REFERENCES [AlignmentClassic].[Alignment]([AlnID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
This is just part of the Multiple Alignment Ontology in OWL DL.
2.4.3 Derived Requirements
This use case leads to the following requirements:
- Map relational data to an RDF schema derived from the relational schema.
- Map from the local ontology to a domain ontology.
- Allow transformation of SPARQL queries over the RDF view into efficient SQL queries.
4 Acknowledgments
The editors gratefully acknowledge contributions from the members of the W3C RDB2RDF Working Group: Marcelo Arenas, Sören Auer, Samir Batla, Richard Cyganiak, Daniel Daniel Miranker, Souripriya Das, Alexander de Leon, Alexander de Leon, Orri Erling, Ahmed Ezzat, Lee Feigenbaum, Angela Fogarolli, Enrico Franconi, Howard Greenblatt, Wolfgang Halb, Harry Halpin, Nuno Lopes, Li Ma, Ashok Malhotra, Ivan Mikhailov, Juan Sequeda, Seema Sundara, Ben Szekely, Edward Thomas, and Boris Villazón-Terrazas.