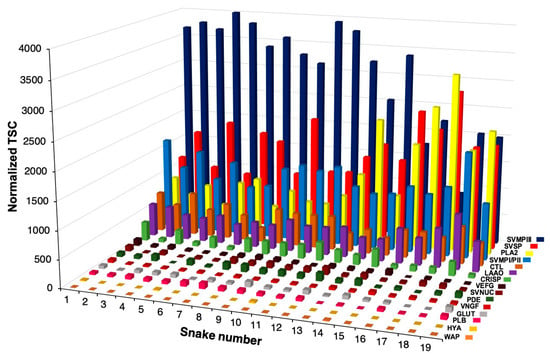
<p>Distribution of the toxin families in the venom of 19 specimens of <span class="html-italic">Bothrops jararacussu</span> identified from proteome analysis. Relative expression is indicated by the normalized Total Spectrum Count (TSC) in each snake. CTL (C-Type Lectin), PLA<sub>2</sub> (Phospholipase A<sub>2</sub>), SVMP classes PI, PII, and PIII (Snake Venom Metalloproteinase), SVSP (Snake Venom Serine Protease), VEGF (Vascular Endothelial Growth Factor), LAAO (L-amino Acid Oxidase), CRISP (Cysteine-rich secretory protein), HYAL (Hyaluronidase), VNGF (Venom Nerve Growth Factor), SVNUC (Snake Venom Nucleotidase), PDE (Venom Phosphodiesterase), PLB (Phospholipase B) and GLUT (Snake Venom Glutaminyl Cyclase). The higher the snake number, the greater the size of the snake.</p> Full article ">Figure 2
<p>Correlation of the main toxin families of <span class="html-italic">Bothrops jararacussu</span> venom with: (<b>A</b>) the individuals’ snout-vent length (SVL), (<b>B</b>) reproductive status, sex, and place of collection. Relative expression is indicated by the normalized Total Spectrum Count (TSC) in each snake. CTL (C-Type Lectin), PLA<sub>2</sub> (Phospholipase A<sub>2</sub>), SVMP classes PI, PII and PIII (Snake Venom Metalloproteinase), SVSP (Snake Venom Serine Protease), LAAO (L-amino Acid Oxidase), CRISP (Cysteine-rich secretory protein). * <span class="html-italic">p</span> < 0.05.</p> Full article ">Figure 3
<p>Distribution of PLA<sub>2</sub> isoforms present in the venoms of 19 individuals from <span class="html-italic">B. jararacussu</span> and functional inferences. Mature deduced protein sequences of PLA<sub>2</sub> were aligned together with sequences of PLA<sub>2</sub>s isolated from other venoms with functions well-established by previous studies using experimental approaches, as follows: a non-toxic phospholipase A2 from the venom of <span class="html-italic">Notechis scutatus scutatus</span>; Bothropstoxin-1 (Q90249.3), Bothropstoxin-2 (P45881) and BthAI (Q8AXY1.1), <span class="html-italic">B. jararacussu</span> venom; acidic PLA<sub>2</sub> (Q2HZ28.1), <span class="html-italic">B. erythromelas</span> venom. <span class="html-italic">Notechis</span> PLA<sub>2</sub> (P08873.1) sequence was used to root the tree. The maximum likelihood phylogenetic tree was generated using RaxML (v8.2.12; Stamatakis 2014). The heatmap on the right indicates the number of normalized Exclusive Unique Spectrum Counts (EUSC) of MS/MS in each venom sample, which are as indicated at the top of the figure. Bootstrap values are described only when greater than 75.</p> Full article ">Figure 4
<p>Distribution of SVMPIII isoforms present in the venoms of 19 individuals from <span class="html-italic">B. jararacussu</span> and functional inferences. Mature deduced protein sequences of SVMPIII were aligned together with sequences of SVMPs isolated from other venoms with functions well-established by previous studies using experimental approaches, as follows: BjussuMP-1P (Q1PHZ4; from <span class="html-italic">B. jararacussu</span> venom; Batroxrhagin, <span class="html-italic">Bothrops atrox</span> venom (ALB00542.1); hemorrhagic factor 3 (HF3), <span class="html-italic">B. jararaca</span> venom (Q98UF9.3); vascular apoptosis-inducing protein 1 (VAP-1), <span class="html-italic">Crotalus atrox</span> venom (Q9DGB9.1); berythractivase, <span class="html-italic">B. erythromelas</span> venom (Q8UVG0.1); VaF1, <span class="html-italic">Vipera ammodytes ammodytes</span> venom (AJC52543). The disintegrin and metalloproteinase domains of <span class="html-italic">Mus musculus</span> ADAM28 (NP034212.1) was used to root the tree. The maximum likelihood phylogenetic tree was generated using RaxML (v8.2.12; Stamatakis 2014). The heatmap on the right indicates the number of normalized Exclusive Unique Spectrum Counts (EUSC) of MS/MS in each venom sample, which are as indicated at the top of the figure. Bootstrap values are described only when greater than 75.</p> Full article ">Figure 5
<p>Comparisons of gene transcription (TMM normalized) rates in the venom gland and protein abundance (normalized EUSC—Exclusive Unique Spectrum Count) in the venom of individual <span class="html-italic">B. jararacussu</span> snakes. Each spot represents detected transcription and translation levels for a specific toxin family from one specimen. Dotted lines indicate a hypothetical correspondence between transcript and protein abundances.</p> Full article ">Figure 6
<p>Comparison between levels of transcripts (TMM normalized) and protein abundance (normalized EUSC—Exclusive Unique Spectrum Count) of the isoforms PLA002, SVMPIII002, and SVMPIII009 in the venoms of <span class="html-italic">B. jararacussu</span> snakes. Dotted lines indicate a hypothetical correspondence between transcript and protein abundances.</p> Full article ">Figure 7
<p>Functional activities of <span class="html-italic">Bothrops jararacussu</span> venom samples. (<b>A</b>)—Metalloproteinase activity (SVMP) was measured by fluorometric assays using the substrate Abz-AGLA-EDDnp. (<b>B</b>)—Phospholipase A<sub>2</sub> (PLA<sub>2</sub>) activity was tested using the 4-nitro-3- [octanoyloxy] benzoic acid substrate. (<b>C</b>)—Procoagulant activity was evaluated by the amount of venom (µg) capable of inducing the coagulation of human citrated plasma in up to 60 s and is represented by the reciprocal Minimum Coagulant Dose (1/MCD). (<b>D</b>)—For hemorrhagic activity, 10 µg of each pool of venoms were injected in the dorsum of mice, and the hemorrhagic lesions were measured three hours after injection. (<b>E</b>)—The myotoxic activity was assayed using 50 µg of pools of venoms injected intramuscularly into the gastrocnemius muscle in Swiss mice, and after 3 h, the sera were assayed for creatine–kinase activity. (<b>F</b>)—For the evaluation of lethal activity, samples containing 200 μg of each venom in a final volume of 200 μL were injected intraperitoneally into groups of four mice. Phosphate-buffered saline (PBS)—was the control in all in vivo experiments. The results are representative of two independent experiments. <b>*</b> <span class="html-italic">p</span> < 0.05.</p> Full article ">